Our Precision Medicine Capabilities
Across all capabilities, PrecisionGo is designed to support complexity rather than simplify it away. Integration, adaptability, and governance are treated as foundational elements, enabling research and clinical work to proceed with greater clarity and confidence. This means fewer disconnected steps, clearer pathways, and infrastructure that is designed to support translation as work evolves. Rather than offering isolated services, PrecisionGo is designed to help complex research and clinical programs function as integrated pathways - and our MRFF-backed infrastructure and expert staff ensure that every step is optimised for speed, quality, and impact.
How PrecisionGo supports integrated precision medicine
Across all capabilities, PrecisionGo is designed to support complexity rather than simplify it away. Integration, adaptability, and governance are treated as foundational elements, enabling research and clinical work to proceed with greater clarity and confidence. This means fewer disconnected steps, clearer pathways, and infrastructure that is designed to support translation as work evolves. Rather than offering isolated services, PrecisionGo is designed to help complex research and clinical programs function as integrated pathways.
Housed at WIMR within Westmead Research Hub, we have the end-to-end facilitaties to provide for your precision medicine needs. The latest cutting-edge technologies, supported by MRFF Grants, means that whether you would like to hand your samples to us and then receive insights from your data, or whether you’d prefer to be involved in any or all of the steps along the way, we’re here to support you in bringing precision medicine to clinical care.
How researchers and clinicians engage with PrecisionGo
PrecisionGo operates through a single, coordinated front door. Instead of needing to approach multiple facilities or teams independently, researchers and clinicians engage with PrecisionGo with one main point of contact. An initial discussion helps clarify the research or clinical question, study context, and intended trajectory, and PrecisionGo then coordinates access across relevant technologies, analysis, and governance. Your user experience has:
one point of entry rather than multiple parallel requests
coordinated planning across facilities and disciplines
support in aligning study design, analysis, and downstream translation
clear oversight and accountability as work progresses
This way, we reduce your administrative burden while ensuring that complex studies are supported coherently from study design through to insight creation.
The PrecisionGo pathway
PrecisionGo supports work across the full precision medicine pathway, from study or clinical question through to analysis and translation. At a high level, this involves:
supporting study design that anticipates downstream needs
coordinating access across multiple modalities and facilities
enabling secure data integration and collaboration
aligning governance, quality, and oversight from the outset.
This approach allows for a single point of contact to a connected pathway from discovery through validation to clinical translation, designed to reduce loss of momentum between research insight and patient impact. In order to reduce fragmentation and support a clearer line of sight from discovery through to impact, we ensure that:
samples, data, and analytical context are carried forward rather than reset
analysis builds on prior work rather than starting again at each stage
clinical relevance is considered early, not introduced late.
Discovery – identifying novel biomarkers
The discovery stage focuses on generating new biological insight from well-characterised patient samples. This typically involves exploration and insight generation, with analysis designed to inform what should be taken forward for further testing:
a small number of blood or tissue samples linked with relevant clinical data
application of advanced genomics and multiomic technologies
integrated data analysis to identify novel biomarkers or biological signals
Screening – validating discoveries at scale
Promising findings from discovery can then move into a screening pathway designed for robustness and validation. This stage focuses on validating biomarkers and assess their consistency, relevance, and potential utility across broader populations:
larger patient cohorts with linked clinical data (which can be sourced through the Westmead Biobank or from recruited patients)
more standardised and automated analytical approaches involving Flow Cytometry, Imaging, Bioresources and Preclinical Imaging, etc.
explicit linkage between laboratory findings and clinical characteristics.
Intentionally coordinated services and technologies
PrecisionGo brings together a deliberately connected ecosystem of technologies and specialist expertise. Capabilities are organised into domains that can be combined as needed, rather than accessed in isolation. These domains include:
Histology – from tissue processing to slide preparation
Genomics – whole genome, targeted, and spatial transcriptomics
Proteomics – protein profiling and biomarker discovery
Cytometry – advanced flow cytometry and cell sorting
Bioresources – animal model supply and husbandry
Preclinical Imaging – in vivo imaging technologies
Imaging – high-resolution microscopy and analysis
Biobanking – ISO9001-aligned biospecimen storage and tracking
Bioinformatics – multi-omics analysis and integrated data interpretation
Integrated precision medicine pipelines
PrecisionGo supports integrated genomics pipelines that are designed to bring multiple layers of biological information together within a single, coherent workflow. Rather than treating each assay or platform as a standalone step, pipelines are designed to align experimental design, data generation, and analysis from the outset.
Current genomics capabilities are organised into three complementary pipelines, each addressing a distinct but related class of research and translational questions. These pipelines are not intended to operate in isolation. They can be used independently or combined within a single study, depending on the research or clinical question. This approach allows studies to evolve as questions develop, while maintaining coherence across modalities, teams, and analysis. Across all three pipelines:
workflows are tailored to study context rather than fixed templates
downstream analysis is considered from the beginning
data is generated with integration and interpretation in mind.
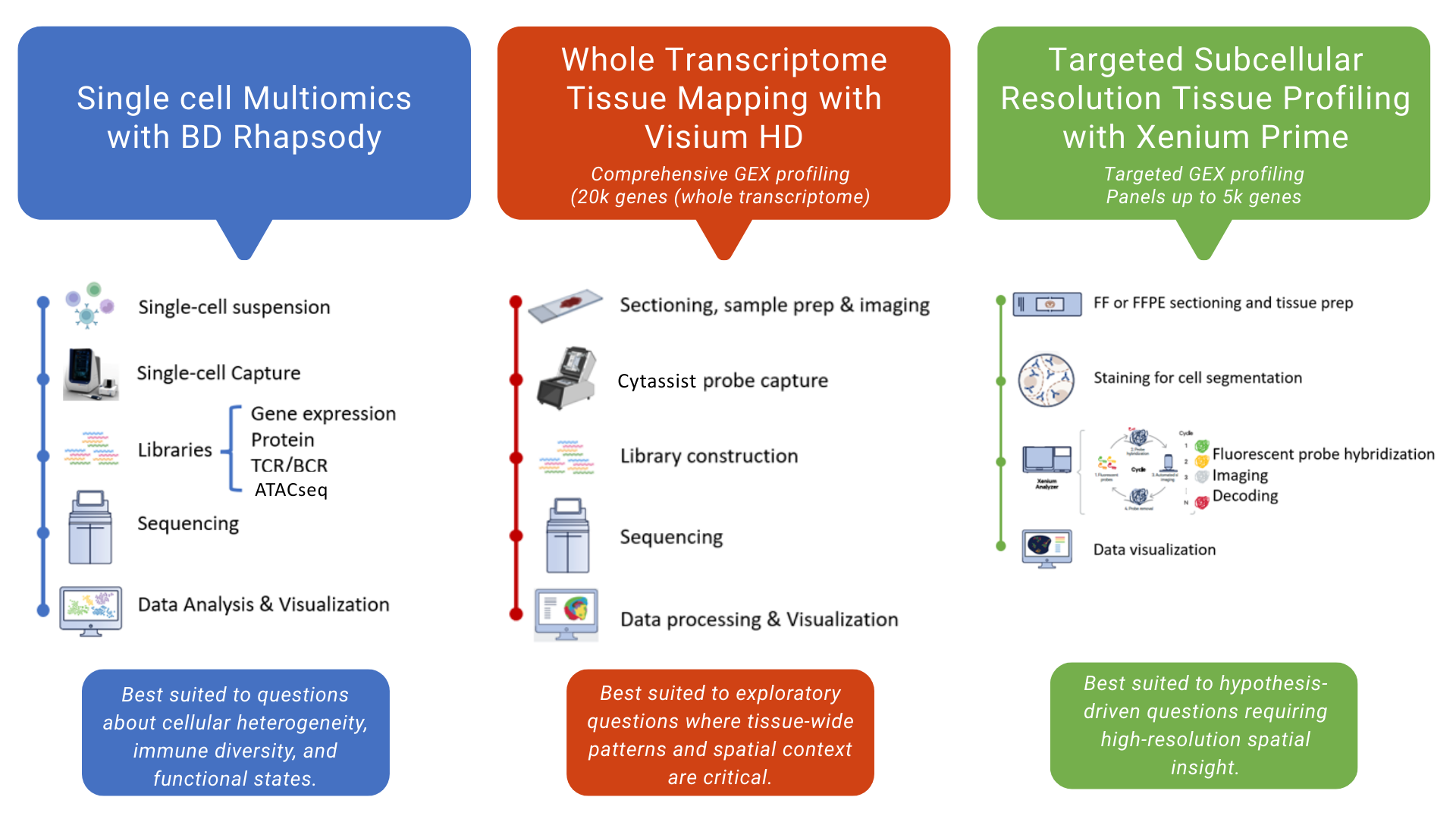
1. Single-cell multiomic profiling
This pipeline enables integrated analysis of individual cells by combining gene expression with additional molecular and cellular features, such as immune receptor diversity and protein expression. It supports studies seeking to:
characterise cellular heterogeneity
link transcriptional states to immune or functional phenotypes
explore clonal diversity and lineage relationships
Through generating multiple data types from the same cells, this pipeline supports richer interpretation and more precise biological insight than single-modality approaches.
2. Whole-transcriptome spatial profiling
This pipeline enables spatially resolved transcriptomics across tissue sections, capturing genome-wide gene expression while preserving tissue architecture. It is suited to questions where spatial context is critical, such as:
tissue organisation and microenvironments
spatial patterns of disease or response
linking molecular profiles to histology
The workflow integrates tissue preparation, transcriptome capture, sequencing, and downstream analysis into a single coordinated pipeline.
3. Targeted spatial transcriptomics
This pipeline provides high-resolution, targeted gene expression profiling at subcellular scale, allowing focused interrogation of specific pathways or cell populations within intact tissue.
It is particularly valuable when:
specific gene panels are of interest
fine spatial resolution is required
analysis needs to be tightly linked to morphology or cell segmentation
Through targeting defined gene sets, this pipeline supports detailed spatial insight while maintaining analytical focus.
The digital platform
The PrecisionGo digital platform functions as connective tissue across the pathway to ensure that insights are generated consistently, efficiently, and with continuity across studies, rather than being rebuilt from scratch each time.
For researchers and clinicians, this means that analytical approaches are informed by prior work, refined over time, and applied with greater confidence. The platform enables bioinformatics teams to reuse and adapt validated analytical modules and workflows developed through previous projects, accelerating analysis while maintaining rigour and traceability. For you, this means:
Faster time to insight: By building on existing, tested analytical approaches rather than reinventing analyses for each study.
Greater consistency and defensibility of results: Through reuse of validated workflows that support reproducibility and clear interpretation.
Continuity across projects and teams: Retaining analytical knowledge and approaches as projects evolve or personnel change, reducing reliance on ad-hoc solutions.
Clearer pathways from data to interpretation: Acting as the analytical bridge between data generated by technologies and the insights used by researchers and clinicians.
The platform is primarily used by bioinformatics teams, so researchers and clinicians are not required to engage with yet another system. Instead, they benefit from more reliable, timely, and interpretable outputs that are better positioned for translation. As PrecisionGo evolves, the platform is designed to support appropriate project visibility and data access, while remaining focused on its core purpose: turning accumulated analysis into shared capability that strengthens current and future work.